Date: Mon Jul 16 2012 - 09:04:17 MDT
Hi Madeleine,
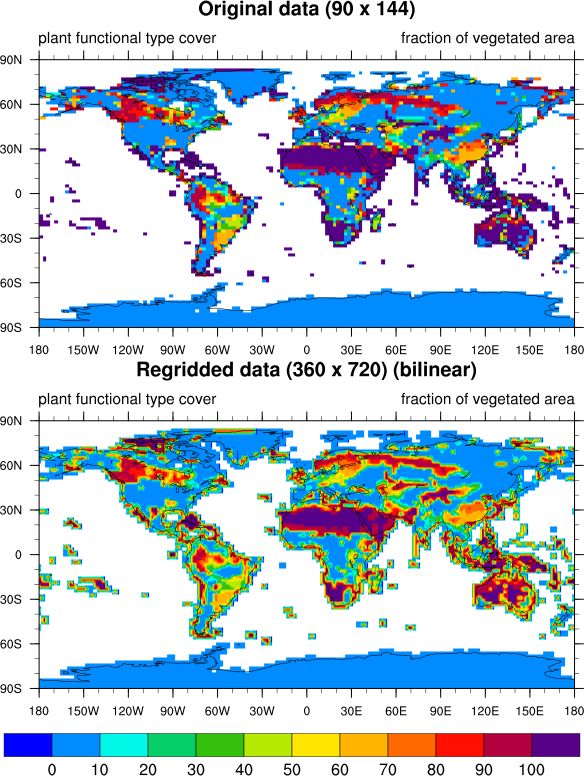
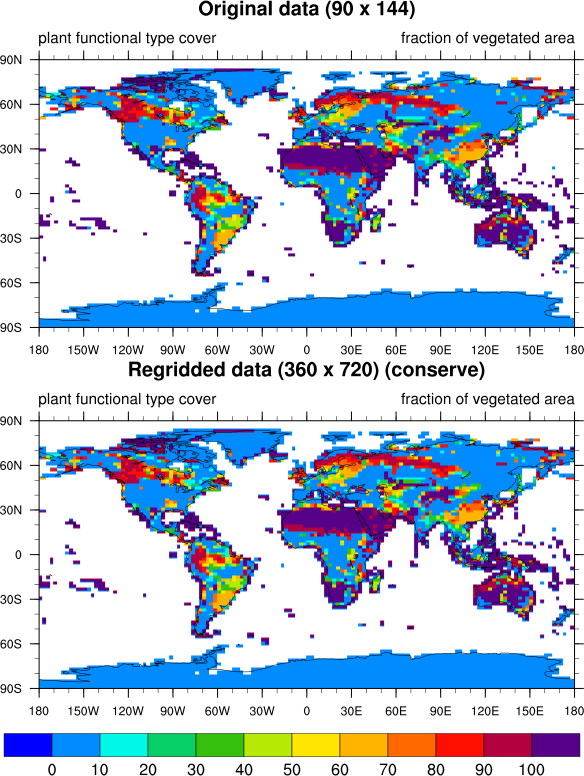
Your question is not a silly one! In fact, I'm going to post it back to ncl-talk, since I should have used a different interpolation method on your variable.
"Bilinear" is just the default regridding method, which I always use for demonstration purposes since it's fast.
But, you're right, the "conserve" method should
be more appropriate for categorical data.
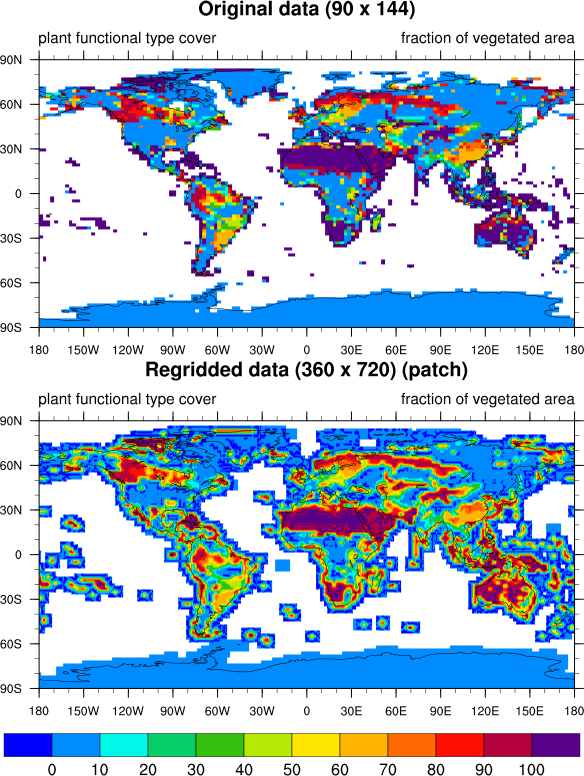
Usually I include the regridding method used in the title of the plot, but I just forgot to do that here.
I've included what it looks like to use all three interpolation methods on your variable. I've also included a new version of your script that allows you to select the interpolation method to use.
The method you use will get put in the title, and it will be used to name the weights file.
--Mary
On Jul 15, 2012, at 1:35 PM, Madeleine Patterson wrote:
> Hi again -sorry for the multiple emails - I didn't want to post this to the list in case it turns out to be a really silly question: but I notice in the output as the script executes, the regridding method is 'bilinear' for the weights - what are these weights?- it's not obvious from the output what the regridding method is for the categorical variable that I want to regrid (fpcgrid, which is percent cover of the plant functional types -i.e. vegetation types), as I assume it must be some other method than 'bilinear' as this is an interpolation method, which is unsuitable for categorical data (isn't it? The UCAR website says it is).
>
> Thanks again,
>
> Madeleine
>
>
>
> On Sat, Jul 14, 2012 at 6:25 AM, Mary Haley <haley@ucar.edu> wrote:
> Hi Madeleine,
>
> I'm posting this back to ncl-talk, so they can see how we resolved this issue.
>
> Please see the attached script, which requires V6.1.0-beta in order to run. This script regrids several variables (I picked them at random, but made the first one "FPCGRID") using the ESMF_regrid function.
>
> I've included the plot of the FPCGRID variable only.
>
> The main regridding function in this script is called "regrid_variable". The first time you regrid something, it generates 3 NetCDF files (a source description file, a destination description file, and a weights file). For regridding other variables from and to the same grid, you don't need to keep regenerating these three files. That's why I have the "SkipSteps" variable. It will skip those three steps for subsequent variables.
>
> In the regrid_variable function, I have to "fix" the latitude values so they are not missing. Otherwise, you will get errors when you try to plot it. Also, at least one of your variables (PFT) had a _FillValue attribute of 9999, which I believe is supposed to be -9999. If you regrid this variable, you will have to fix the _FillValue (I have a check for this in the attached script).
>
> --Mary
>
>
>
> On Jul 9, 2012, at 10:10 AM, Mary Haley wrote:
>
> > Madeleine,
> >
> > I don't understand something about your script. You are setting clat and clon to scalar values, and then using these in the grid2triple routine. Are you really trying to regrid only one value at a time?
> >
> > grid2triple is expecting the first two arguments to be arrays of lat,llon values of size nlat and nlon. The third argument to grid2triple, then, should be an nlat x long array whose coordinates are represented by the lat and lon arrays.
> >
> > You are giving it a 2D array to regrid, but only one longitude and one latitude value. That's why it's giving you an error.
> >
> > I might be able to help you with the regridding issue, using our new ESMF regridding capabilities. Can you give me your full script and data, and a detailed explanation of what you are trying to regrid? Do you have V6.1.0-beta installed?
> >
> > --Mary
> >
> >
> > On Jul 8, 2012, at 6:29 PM, Madeleine Patterson wrote:
> >
> >> Hi,
> >>
> >> I tried respecifying it to have 3 dimensions (pctpftN[*][*][*]), took out the do loops, but am still getting error messages - it seems that the grid2triple function cant handle 3 dimensions (which I guess should be apparent from it's name...)
> >>
> >> fatal:Number of dimensions in parameter (2) of (grid2triple) is (3), (2) dimensions were expected
> >> fatal:Execute: Error occurred at or near line 27 in file changedims.ncl
> >>
> >> ...If I use do loops to try to use grid2triple with a 3rd dimension, I get an error:
> >>
> >> fatal:grid2triple: The last input array must be dimensioned ny x mx, where ny is the length of y, and mx is the length of x
> >>
> >> So, I can't get it to work.... seems I may not be able to regrid my categorical data this way...
> >>
> >>
> >>
> >>
> >> On Sat, Jul 7, 2012 at 5:46 AM, Hobbs, Will R (3244-Affiliate) <William.R.Hobbs@jpl.nasa.gov> wrote:
> >> Madeleine
> >>
> >> In your argument for the function regrid_cat(), you specify the input variable pctpftN as having two dimensions (i.e. pctpftN[*][*]).
> >>
> >> Hope that helps
> >>
> >> Will
> >>
> >> **********************************************************
> >> Will Hobbs, Ph.D. William.R.Hobbs@jpl.nasa.gov
> >> Jet Propulsion Laboratory
> >> 4800 Oak Grove Dr. office: 300-324a
> >> M/S 300-323 phone: (818) 354-0466
> >> Pasadena, CA 91109 fax: (818) 354-0966
> >> **********************************************************
> >>
> >> From: Madeleine Patterson <madeleine.patterson77@gmail.com>
> >> Date: Sat, 7 Jul 2012 05:40:57 +1000
> >> To: ncl-talk <ncl-talk@ucar.edu>
> >>
> >> Subject: Re: Regridding from low to high resolution ?
> >>
> >> Hi,
> >> I am still trying to implement some script ideas that Mary Haley suggested I try in order to regrid categorical data from low to high resolution, but I cannot get the grid2triple() function -which I'm using to create a regrid function, let alone the actual regrid function - to work with 3 dimensions of data -even with do loops... I keep getting error messages telling me it's expecting 2 dims when I'm giving it 3:
> >>
> >> fatal:Number of subscripts do not match number of dimensions of variable,(3) Subscripts used, (2) Subscripts expected
> >> fatal:Execute: Error occurred at or near line 23 in file changedims.ncl
> >> fatal:Execute: Error occurred at or near line 75 in file changedims.ncl
> >>
> >> I am wondering if I am implementing the do loop with the wrong syntax, or if there is something else wrong with my script (below)?
> >> Can anyone suggest a way to get grid2triple to work with a 3D input dataset?
> >>
> >> Thanks for your advice...
> >>
> >> MP
> >>
> >> ********************************************************************************
> >> load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_code.ncl"
> >> load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_csm.ncl"
> >> load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/contributed.ncl"
> >> load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/shea_util.ncl"
> >>
> >> undef("regrid_cat")
> >> function regrid_cat(clat[*],clon[*],pctpftN[*][*],tlat[*],tlon[*]) ; categorical grid and target grid
> >> local testd, pctpftNnew
> >>
> >> begin
> >>
> >> do i=1,10
> >> testd=grid2triple(clon,clat,pctpftN(i,:,:)) ; testd(3,ld)
> >> end do
> >>
> >> pctpftNnew = triple2grid(testd(0,:),testd(1,:),testd(2,:),tlon,tlat,False)
> >> copy_VarAtts(pctpftN,pctpftNnew) ;contributed.ncl
> >> return(pctpftNnew)
> >>
> >> end
> >> ;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;
> >> ; main code
> >> ;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;;
> >> begin
> >>
> >> in1 = addfile("pctpft.nc","r")
> >> pctpft = in1->PCTPFT
> >> pctpftN = pctpft(0,:,:,:)
> >>
> >> printVarSummary(pctpftN)
> >>
> >> clat=90
> >> clon=144
> >> tlat=360
> >> tlon=720
> >>
> >> ntim =dimsizes(pctpftN(:,0,0))
> >> nlat =dimsizes(tlat)
> >> nlon = dimsizes(tlon)
> >>
> >> pctpftNnew=new((/ntim,nlat,nlon/),typeof(pctpftN))
> >> ;pctpft_regrid=new((/ntim,nlat,nlon/),typeof(pctpftN))
> >>
> >> do i=0,ntim-1
> >>
> >> pctpftNnew(i,:,:) = regrid_cat(clat,clon,pctpftN(i,:,:),tlat,tlon) ; categorical grid and target grid
> >>
> >> end do
> >>
> >> end
> >> ********************************************************************************
> >>
> >> On Tue, Jun 19, 2012 at 2:33 AM, Mary Haley <haley@ucar.edu> wrote:
> >> Hi Madeleine,
> >>
> >> You will need to add a "do" loop and call grid2triple for each element of that third dimension. I'm not sure why grid2triple doesn't allow for multiple leftmost dimensions. I'll look into this.
> >>
> >> Meanwhile, maybe something like this:
> >>
> >> ntim = dimsizes(pctpft(:,0,0))
> >> nlat = dimsizes(tlat)
> >> nlon = dimsizes(tlon)
> >>
> >> xregrid = new( (/ntim,nlat,nlon/), typeof(pctpft))
> >>
> >> do i=0,ntim-1
> >> xregrid(i,:,:) = regrid_cat(clon,clat,pctpft(i,:,:),tlat,tlon)
> >> end do
> >>
> >> I don't know what "regrid_cat" looks like, so I'm not sure this is the correct syntax. Perhaps the creation of "xregrid" and the "do" loop should be moved into "regrid_cat".
> >>
> >> --Mary
> >>
> >> On Jun 15, 2012, at 10:23 AM, Madeleine Patterson wrote:
> >>
> >>> Hi Mary,
> >>>
> >>> Thanks for your response - I am trying to get the regridding function I created to work; I've tried your code but I get an error message:
> >>>
> >>> fatal: number of dimensions in parameter (2) or (regrid_cat) is (3), (2) dimensions were expected
> >>>
> >>> So I tried putting the regrid function into a do loop so I can loop over all 10 pfts...
> >>>
> >>> do i=1,10
> >>> xregrid = regrid_cat(clon,clat,pctpft(i,:,:),tlat,tlon) ;;;; (this may well be incorrect syntax)
> >>> end do
> >>>
> >>> but then I get the error message :
> >>>
> >>> fatal: grid2triple : the last input array must be dimensioned ny x mx where ny is the length of y and mx is the length of x
> >>>
> >>> - so in my function definition testd=grid2triple(clon,clat,pctpft), it's saying pctpft must be 2 dimensional, not 3 dimensional (which it is).
> >>>
> >>> How to get around this? Would I have to read in each pft as one 2d variable, perform the regridding then reconstruct the 3d netcdf file, or is there an easier solution?
> >>>
> >>> thanks,
> >>>
> >>> M.P.
> >>
> >>
> >>
> >> _______________________________________________ ncl-talk mailing list List instructions, subscriber options, unsubscribe: http://mailman.ucar.edu/mailman/listinfo/ncl-talk
> >>
> >> _______________________________________________
> >> ncl-talk mailing list
> >> List instructions, subscriber options, unsubscribe:
> >> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
> >
>
>
>
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk