Date: Fri Aug 17 2012 - 09:39:27 MDT
Hi NCL,
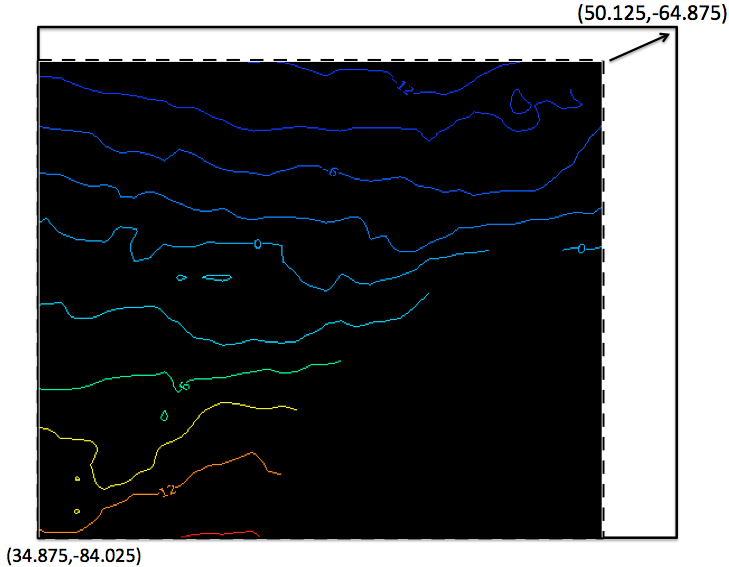
I am asking if there is a function in NCL like the GIS buffering, e.g., I
have a data(in the dash box show below), What I want to do is like this:
Buffer(or interpolate/regridding) the dashed box data to the solid
box(where the space between the dashed box and solid box was no data there)
but keep the original cell value in the dashed box no change.
[image: Inline image 2]
Have someone have done similar thing before. I will be appreciated if you
can give me suggestions and advice.
Thanks,
Ping
On Mon, Aug 13, 2012 at 11:47 PM, Dennis Shea <shea@ucar.edu> wrote:
> The error message is telling you exactly what the error is.
>
> V = cru1990->precipitation ==> V(12,360,720)
>
> Your new lat/lon are (306,384)
>
>
> newlon = fspan(-84.025,-64.875,384)
> newlat = fspan(34.875,50.125,306)
>
> Hence,
>
> linint2_Wrap(V&longitude,V&**latitude,V,True,newlon,newlat,**0)
>
> would be result in a (12,306,384) array.
>
> So, you are trying to place a (12,306,384) array into a (12,360,720).
> Like the error message says, "Dimension sizes of left hand side and
>
> right hand side of assignment do not match."
>
> Currently, NCL does not allow this. (It will in a future release.)
>
> Change to:
>
> Vnew = linint2_Wrap(V&longitude,V&**latitude,V,True,newlon,newlat,**0)
> printVarSummary(Vnew)
>
>
>
>
> On 8/13/12 9:28 PM, Ping Yang wrote:
>
>> Hi Adam,
>>
>> I got a problem with the regridding using linint2_Wrap(it was working
>> for the daily data) with a monthly data,
>> the is an error showed:
>> fatal:Dimension sizes of left hand side and right hand side of
>> assignment do not match.
>> I ncdump -c the data file:
>> netcdf Global_Precipitation_CRU21_**30min_mT1980 {
>> dimensions:
>> latitude = 360 ;
>> longitude = 720 ;
>> bnds = 2 ;
>> time = UNLIMITED ; // (12 currently)
>> variables:
>> double latitude(latitude) ;
>> latitude:long_name = "Latitude" ;
>> latitude:standard_name = "latitude_north" ;
>> latitude:units = "degrees_north" ;
>> latitude:valid_range = -90., 90. ;
>> latitude:actual_range = -90., 90. ;
>> latitude:bounds = "longitude_bnds" ;
>> latitude:axis = "y" ;
>> double longitude(longitude) ;
>> longitude:long_name = "Longitude" ;
>> longitude:standard_name = "longitude_east" ;
>> longitude:units = "degrees_east" ;
>> longitude:valid_range = -180., 180. ;
>> longitude:actual_range = -180., 180. ;
>> longitude:axis = "x" ;
>> double latitude_bnds(latitude, bnds) ;
>> double longitude_bnds(longitude, bnds) ;
>> int time(time) ;
>> time:long_name = "Time" ;
>> time:standard_name = "time" ;
>> time:units = "months since 1980-01-01 00:00" ;
>> time:axis = "t" ;
>> time:actual_range = 0, 11 ;
>> double minimum(time) ;
>> double maximum(time) ;
>> double average(time) ;
>> double stddev(time) ;
>> float precipitation(time, latitude, longitude) ;
>> precipitation:long_name = "Glo_CRU21_Precipitation_**mTS1980_30min" ;
>> precipitation:standard_name = "Precipitation" ;
>> precipitation:var_desc = "Precipitation" ;
>> precipitation:missing_value = -999.f ;
>> precipitation:_FillValue = -999.f ;
>> precipitation:scale_factor = 1.f ;
>> precipitation:add_offset = 0.f ;
>> precipitation:actual_range = 0.f, 1832.9f ;
>>
>> // global attributes:
>> :Conventions = "CF-1.2" ;
>> :title = "Glo_CRU21_Precipitation_**mTS1980_30min" ;
>> :data_type = "continuous" ;
>> :domain = "Global" ;
>> :subject = "precipitation" ;
>> :references = ;
>> :institution = ;
>> :source = ;
>> :comments = ;
>> data:
>>
>> latitude = -89.75, -89.25, -88.75, -88.25, -87.75, -87.25, -86.75,
>> -86.25,
>> -85.75, -85.25, -84.75, -84.25, -83.75, -83.25, -82.75, -82.25,
>> -81.75,
>> -81.25, -80.75, -80.25, -79.75, -79.25, -78.75, -78.25, -77.75,
>> -77.25,
>> ......................
>> 77.25, 77.75, 78.25, 78.75, 79.25, 79.75, 80.25, 80.75, 81.25, 81.75,
>> 82.25, 82.75, 83.25, 83.75, 84.25, 84.75, 85.25, 85.75, 86.25, 86.75,
>> 87.25, 87.75, 88.25, 88.75, 89.25, 89.75 ;
>>
>> longitude = -179.75, -179.25, -178.75, -178.25, -177.75, -177.25,
>> -176.75,
>> -176.25, -175.75, -175.25, -174.75, -174.25, -173.75, -173.25,
>> -172.75,
>> ..........................
>> 168.75, 169.25, 169.75, 170.25, 170.75, 171.25, 171.75, 172.25,
>> 172.75,
>> 173.25, 173.75, 174.25, 174.75, 175.25, 175.75, 176.25, 176.75,
>> 177.25,
>> 177.75, 178.25, 178.75, 179.25, 179.75 ;
>>
>> time = 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11 ;
>> }
>> the longitude and latitude is increasing monotonically
>> and I used the following code to do the regridding:
>>
>> cru1990=addfile(idCRU+"Global_**Precipitation_CRU21_30min_**
>> mT1980.nc","r");
>>
>> V=cru1990->precipitation(:,:,:**)
>>
>> newlon = fspan(-84.025,-64.875,384)
>>
>> newlat = fspan(34.875,50.125,306)
>>
>> ;**************************************************
>>
>> ; interpolate to new grid use linint2_Wrap function
>>
>> ;*************************************************
>>
>> V=linint2_Wrap(V&longitude,V&**latitude,V,True,newlon,newlat,**0)
>>
>>
>> and there was an error:
>>
>> fatal:Dimension sizes of left hand side and right hand side of
>> assignment do not match.
>>
>> Could anyone can help me on this?(Please let me know if there is a place
>> to upload my sample file).
>>
>> Looking forward to hearing from you.
>>
>> Regards,
>>
>> Ping
>> On Thu, May 17, 2012 at 12:45 PM, Adam Phillips <asphilli@ucar.edu
>> <mailto:asphilli@ucar.edu>> wrote:
>>
>> Hi Ping,
>> I think you are on the right track, but you do not need to subset the
>> input array. (I am guessing that you are getting null values around
>> the
>> perimeter of your new grid.) When it comes to regridding one needs
>> to make
>> sure that the input latitudes and longitudes and the output
>> latitudes and
>> longitudes are in the same range, and flow in the same direction. You
>> state you want your grid on negative longitudes, which is fine, but
>> that
>> means your input longitudes need to span the same range as well. So, I
>> would recommend reading in the input data in its' entirety, flipping
>> the
>> latitudes, then use lonFlip to convert the longitudes from 0:360 to
>> -180:180:
>>
>> pres=in->pres(:,::-1,:) ; ::-1 = flip latitudes
>> pres = lonFlip(pres)
>> printVarSummary(pres) ; lats should run from S->N, and lons from
>> -180:180
>>
>> newlon = fspan(-84.025,-64.875,383)
>> newlat = fspan(34.875,50.125,305)
>> newlat@units = "degrees_north"
>> newlon@units = "degrees_east"
>> newsst=linint2_Wrap(pres&lon,**pres&lat,pres,True,newlon,**newlat,0)
>>
>> That should be it. If that doesn't work or if you have further
>> questions
>> please respond to ncl-talk.
>> Best regards,
>> Adam
>>
>>
>> > Dear NCL user,
>> >
>> > I am doing interpolation from the PSD data(daily T62 Gaussian grid
>> > (192x94)
>> > 88.542N - 88.542S, 0E - 358.125E) to a 3 minute*3minute lon/lat
>> regular
>> > grid, I have encountered an issue with the bilinear interpolation
>> with the
>> > linint2_Wrap function (Interpolates from a rectilinear grid to
>> another
>> > rectilinear grid using bilinear interpolation, and retains
>> metadata)with
>> > NCL, because I've got invalid values(some of of the value are
>> null). I
>> > used
>> > this code (I used a NCEP/NCAR reanalysis data, which is available
>> by this
>> > link
>> >
>> ftp://ftp.cdc.noaa.gov/**Datasets/ncep.reanalysis.**
>> dailyavgs/surface_gauss/pres.**sfc.gauss.2006.nc<ftp://ftp.cdc.noaa.gov/Datasets/ncep.reanalysis.dailyavgs/surface_gauss/pres.sfc.gauss.2006.nc>
>> )
>> > :
>> >
>> > ;***************************************************
>> > ; interpolation.ncl
>> > ;
>> > ; Concepts illustrated:
>> > ; - Interpolating from one grid to another using bilinear
>> interpolation
>> > ; use linint2_Wrap function for the retrieving all the metadata
>> > ;create a new file to include the interpolated variable
>> > ;
>> > ;**************************************************
>> > load "/usr/local/lib/ncarg/**nclscripts/csm/contributed.**ncl"
>> > ;load "$NCARG_ROOT/lib/ncarg/**nclscripts/csm/contributed.**ncl"
>> > ;change $NCARG_ROOT to /usr/local directly.
>> > ;**************************************************
>> > begin
>> > ;open file to write variable to(a new netCDF file)
>> > out=addfile("pres.sfc.gauss.**2006.interpolated.nc<http://pres.sfc.gauss.2006.interpolated.nc>
>> <http://pres.sfc.gauss.2006.**interpolated.nc<http://pres.sfc.gauss.2006.interpolated.nc>
>> >","c")
>>
>> > ;**************************************************
>> > ; read in netCDF file
>> > ;**************************************************
>> > in = addfile("pres.sfc.gauss.2006.**nc<http://pres.sfc.gauss.2006.nc>
>> <http://pres.sfc.gauss.2006.nc**>","r")
>>
>> > ;**************************************************
>> > ; read in pres
>> > ;*************************************************
>> > ;Use a subscript for change the dimension of pressure and also
>> subset the
>> > variable
>> > pres=in->pres(time|:,{lat|34.**875:50.125},{lon|95.975:115.**
>> 125})
>> > ;so the newPres is with the latitude increasing
>> > ;*************************************************************
>> > ;newlon = fspan(-84.025,-64.875,383)
>> > newlon = fspan(95.975,115.125,383);**convert to a (0,360) instead
>> of
>> > (-180,+180)
>> > newlat = fspan(34.875,50.125,305)
>> > newlat@units = "degrees_north"
>> > newlon@units = "degrees_east"
>> > ;**************************************************
>> > ; interpolate to new grid use linint2_Wrap function
>> > ;*************************************************
>> > newsst=linint2_Wrap(pres&lon,**pres&lat,pres,True,newlon,**
>> newlat,0)
>> > ;*************output variable to new file****************
>> > out->pres=newsst
>> > end
>> >
>> > Actually I need the domain range is from lon(-84.025,-64.875,383),
>> > lat(34.875,50.125,305) in the lon/lat regular grid, Can I ask you
>> what
>> > will
>> > be the correct range in the Gaussian grid lon 95.975:115.125? I
>> tried
>> > several times, it seems that I am confused by the translation,
>> >
>> > Am I use the right function for this interpolation? I will be
>> appreciated
>> > if you can point out where is the problem ?
>> >
>> > Best Regards,
>> >
>> > Ping
>> >
>> > --
>> > Ping Yang, Ph.D.
>> > CUNY Environmental Crossroads Initiative
>> > Marshak Science Building - Suite 925
>> > The City College of New York - CUNY
>> > 160 Convent Avenue, New York NY 10031
>> > Phone: 212-650-5769
>> > Fax: 212-650-7064
>> > ______________________________**_________________
>> > ncl-talk mailing list
>> > List instructions, subscriber options, unsubscribe:
>> > http://mailman.ucar.edu/**mailman/listinfo/ncl-talk<http://mailman.ucar.edu/mailman/listinfo/ncl-talk>
>> >
>>
>>
>> ______________________________**____________________
>> Adam Phillips asphilli@ucar.edu <mailto:asphilli@ucar.edu>
>>
>> NCAR/Climate and Global Dynamics Division
>> P.O. Box 3000 (303) 497-1726 <tel:%28303%29%20497-1726>
>>
>> Boulder, CO 80307-3000
>> http://www.cgd.ucar.edu/cas/**asphilli<http://www.cgd.ucar.edu/cas/asphilli>
>>
>>
>>
>>
>>
>>
>>
>> ______________________________**_________________
>> ncl-talk mailing list
>> List instructions, subscriber options, unsubscribe:
>> http://mailman.ucar.edu/**mailman/listinfo/ncl-talk<http://mailman.ucar.edu/mailman/listinfo/ncl-talk>
>>
>>
-- Ping Yang, Ph.D. CUNY Environmental Crossroads Initiative Marshak Science Building - Suite 925 The City College of New York - CUNY 160 Convent Avenue, New York NY 10031 Phone: 212-650-5769 Fax: 212-650-7064
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk