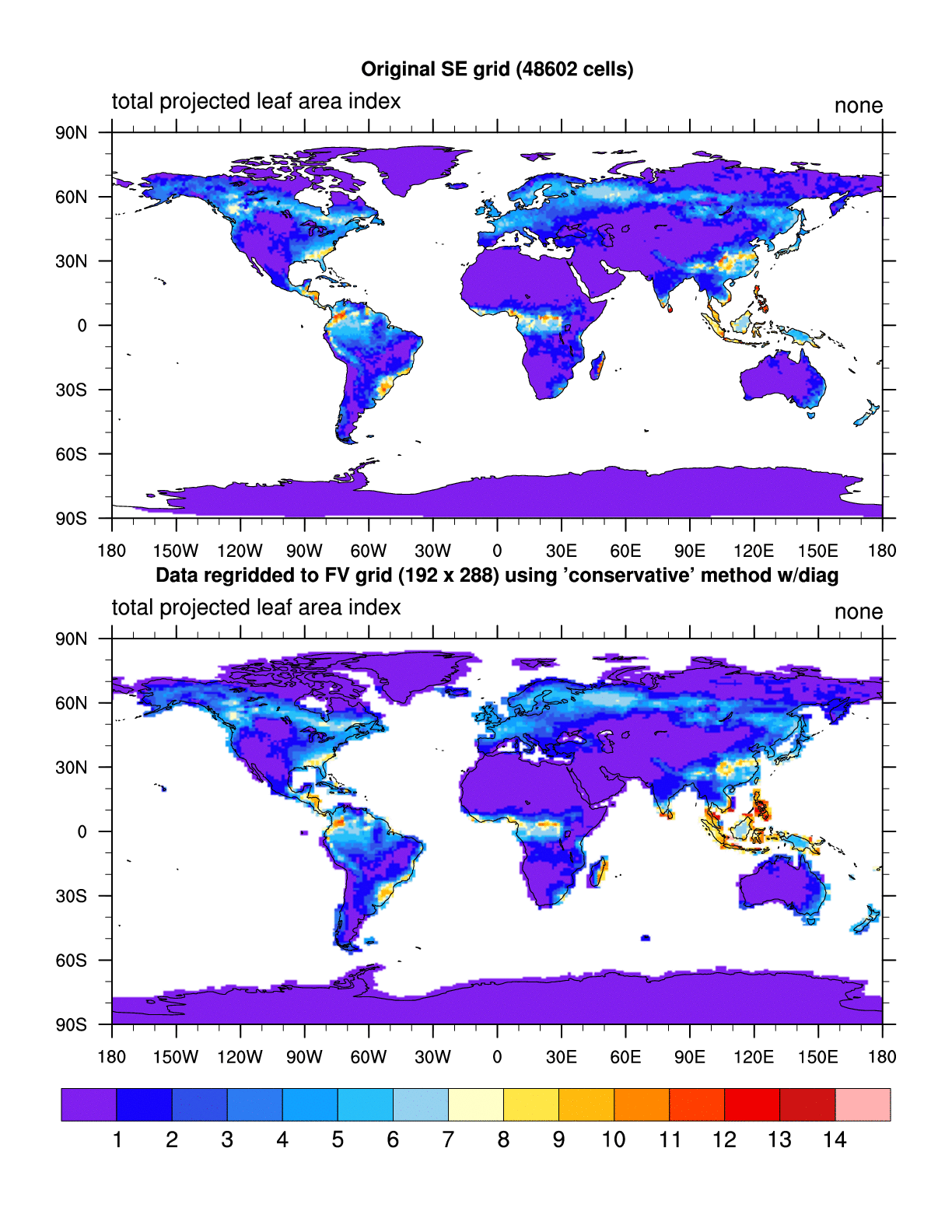
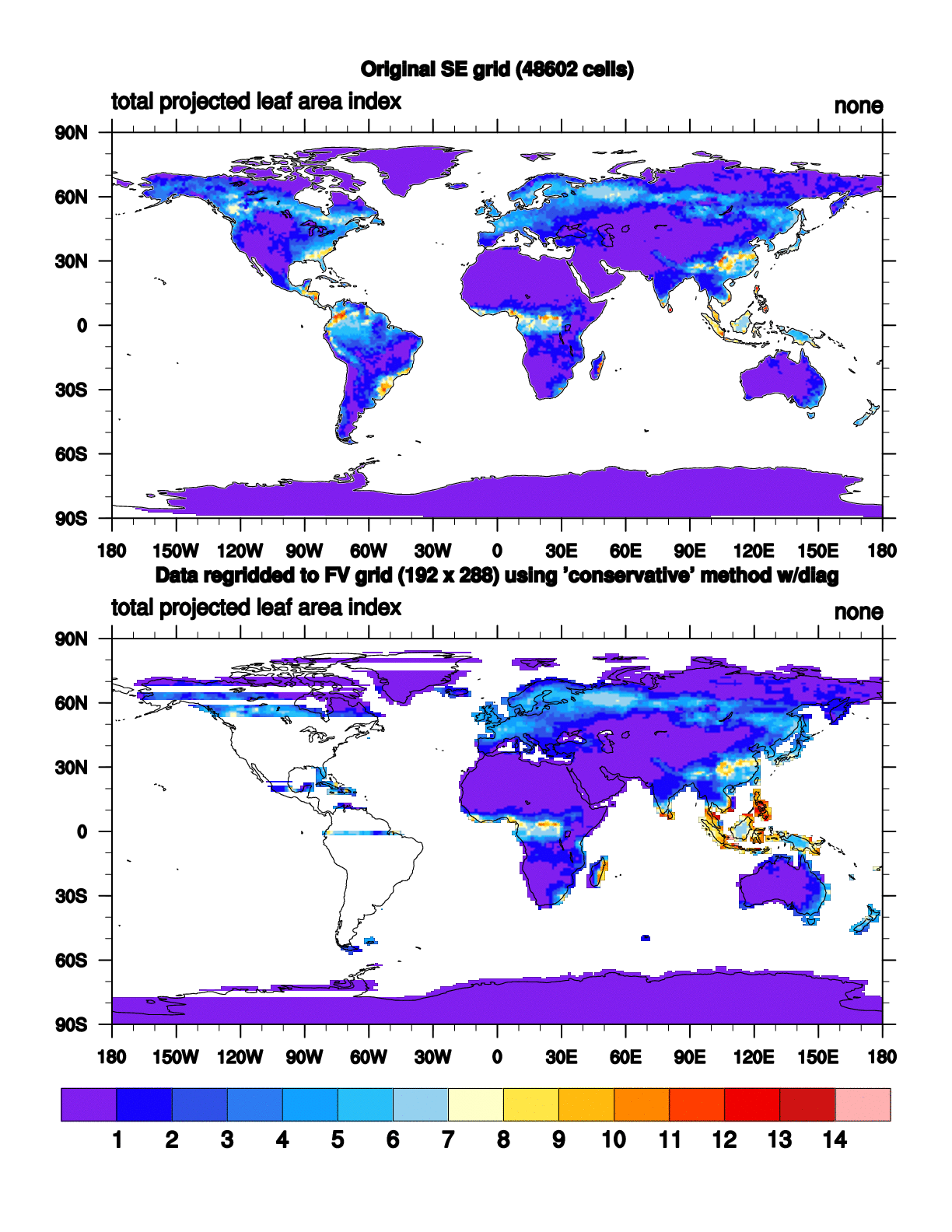
Date: Fri Nov 01 2013 - 13:57:45 MDT
I have a NCL script (attached: plot_SE_FV.ncl, modified from NCL
regridding page) that produces a paneled contour plot.
When I run the script with NCL6.1.2, it runs with no errors and the plot
looks fine (attached: SE_NE30_to_FV_192x288.conserve.NCL6.1.2.gif)
When I run the script with NCL6.2.0, it runs with no errors but the plot
has some strange striping in it (attached:
SE_NE30_to_FV_192x288.conserve.NCL6.2.0.gif).
I converted these from postscript to gif to save space, but they look
the same in ghostview.
Oddly, when I print both postscript files to a color printer they both
look fine.
I would be fine with using NCL6.1.2, but I found that I need NCL6.2.0 to
do regridding properly and I have an integrated script that does
regridding and plotting together.
I am on NCAR yellowstone.
For NCL6.2.0: ncl -V : 6.2.0-01Nov2013_0234
For NCL6.1.2: ncl -V : 6.1.2
Thanks,
Keith
;----------------------------------------------------------------------
; Original code written by Dennis Shea and modeified by Sheri Mickelson
; March 2013 then by Keith Oleson October 2013
;----------------------------------------------------------------------
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_code.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_csm.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/contributed.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/esmf/ESMF_regridding.ncl"
begin
;---Specify remap method
remap_method = "conserve" ; "bilinear" , "patch", "conserve"
;---Specify source SE grid resolution
SE_res = "ne30"
;---Specify name of destination grid
DstGridName = "FV_192x288"
;---Sample plot options
pltDir = "./"
pltType = "ps"
pltName = "SE_"+str_upper(SE_res)+"_to_"+DstGridName
;---Data file containing source grid (sample)
SE_dir = "/glade/scratch/oleson/ANALYSIS/cam5clm45bgc_ne30cesm12rel06_2000b/"
SE_file = "SE_NE30_cam5clm45bgc_ne30cesm12rel06_2000b_ANN_climo.nc"
sfile = addfile(SE_dir + SE_file,"r")
;---Data file containing destination grid
FV_dir = "/glade/scratch/oleson/ANALYSIS/cam5clm45bgc_ne30cesm12rel06_2000b/"
FV_file = "FV_192x288_cam5clm45bgc_ne30cesm12rel06_2000b_ANN_climo_diag_conserve.nc"
dfile = addfile(FV_dir+FV_file,"r")
;---Get SE variable
var_name = "TLAI"
var_in = sfile->$var_name$
src_lat = sfile->lat
src_lon = sfile->lon
;---Get FV variable
var_regrid = dfile->$var_name$
dst_lat = dfile->lat
dst_lon = dfile->lon
;+++
; End user input
;+++
;----------------------------------------------------------------------
; Plotting section
;
; This section creates filled contour plots of both the original
; data and the regridded data, and panels them.
;----------------------------------------------------------------------
dims_in = dimsizes(var_in)
print(dims_in)
rank_in = dimsizes(dims_in)
ntim = dims_in(0)
pltName = pltName+"."+remap_method
pltPath = pltDir+pltName
wks = gsn_open_wks(pltType,pltPath)
;---Resources to share between both plots
res = True ; Plot modes desired.
res@gsnDraw = False
res@gsnFrame = False
res@gsnMaximize = True ; Maximize plot
res@cnFillOn = True ; color plot desired
res@cnLinesOn = False ; turn off contour lines
res@cnLineLabelsOn = False ; turn off contour labels
res@cnFillMode = "RasterFill" ; turn raster on
res@lbLabelBarOn = False ; Will turn on in panel later
res@mpFillOn = False
res@gsnLeftString = var_name ; long_name is too long!
res@gsnLeftString = var_in@long_name
res@gsnCenterString = " "
;---Resources for plotting regridded data
res@gsnAddCyclic = True
dims = tostring(dimsizes(var_regrid))
rank = dimsizes(dims)
res@tiMainFontHeightF = 0.015
res@tiMainString = "Data regridded to FV grid (" + \
str_join(dims(rank-2:)," x ") + ") using '" + \
"conservative" + "' method" + " w/diag"
res@cnLevelSelectionMode = "ExplicitLevels"
;TLAI
res@cnLevels = (/1,2,3,4,5,6,7,8,9,10,11,12,13,14/)
plot_regrid = gsn_csm_contour_map(wks,var_regrid(0,:,:),res)
;---Resources for plotting original (source) data
res@sfXArray = src_lon
res@sfYArray = src_lat
res@gsnAddCyclic = False
; res@trGridType = "TriangularMesh"
res@tiMainString = "Original SE grid (" + dims_in(1) +" cells)"
res@mpFillOn = True
res@mpOceanFillColor = "white"
res@mpLandFillColor = "transparent"
res@mpFillDrawOrder = "postdraw"
plot_orig = gsn_csm_contour_map(wks,var_in(0,:),res)
;---Draw both plots in a panel
pres = True
pres@gsnMaximize = True
pres@gsnPanelLabelBar = True
gsn_panel(wks,(/plot_orig,plot_regrid/),(/2,1/),pres)
end
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk