Date: Wed Apr 14 2010 - 11:30:53 MDT
As Adam indicated you may be running out of memory.
===
I did the following on a local linux box ...
Try
%> ncl_filedump MOD29E1D.A2009340.005.2009341094922.hdf | less
then
%> ncl_filedump MOD29E1D.A2009340.005.2009341094922.hdf.hdfeos | less
From the .hdf ... no lat/lon on file ... just 'cartesian' distances
short Ice_Surface_Temperature_NP ( XDim_MOD_Grid_Seaice_4km_North,
YDim_MOD_Grid_Seaice_4km_North )
long_name : Estimated sea ice surface temperature 4 km North
Pole grid
units : degree_Kelvin
format : f4.1
coordsys : cartesian
valid_range : ( 22320, 31320 )
_FillValue : 7
missing_value : 0x00
scale_factor : 0.01
scale_factor_err : 0
add_offset : 0
add_offset_err : 0
calibrated_nt : 23
Key : 0.0=missing data, 1.0=no decision, 5.0=non-production
mask, 7.0=tile fill, 8.0=no input tile expected, 25.0=land, 37.0=inland
water, 50.0=cloud
hdf_name : Ice_Surface_Temperature_NP
---
hdfeos: .... NCL *creates* new latitude/longitude variables, that
facilitate plotting & processing
short Ice_Surface_Temperature_NP_MOD_Grid_Seaice_4km_North (
XDim_MOD_Grid_Seaice_4km_North, YDim_MOD_Grid_Seaice_4km_North )
coordinates : GridLat_MOD_Grid_Seaice_4km_North,
GridLon_MOD_Grid_Seaice_4km_North
hdfeos_name : Ice_Surface_Temperature_NP
projection : Lambert Azimuthal Equal Area
unsigned : True
Note the "unsigned"
Unfortunately, the *important* attributes: Key, scale_factor,
add_offset, _FillValue, etc are not transferred.
The LAT/LON generated are
double GridLat_MOD_Grid_Seaice_4km_South (
YDim_MOD_Grid_Seaice_4km_South, XDim_MOD_Grid_Seaice_4km_South )
projection : Lambert Azimuthal Equal Area
corners : ( 1e+51, 1e+51, 1e+51, 1e+51 )
There is no indication that values of 1e51 are _FillValue.
Common sense but ....
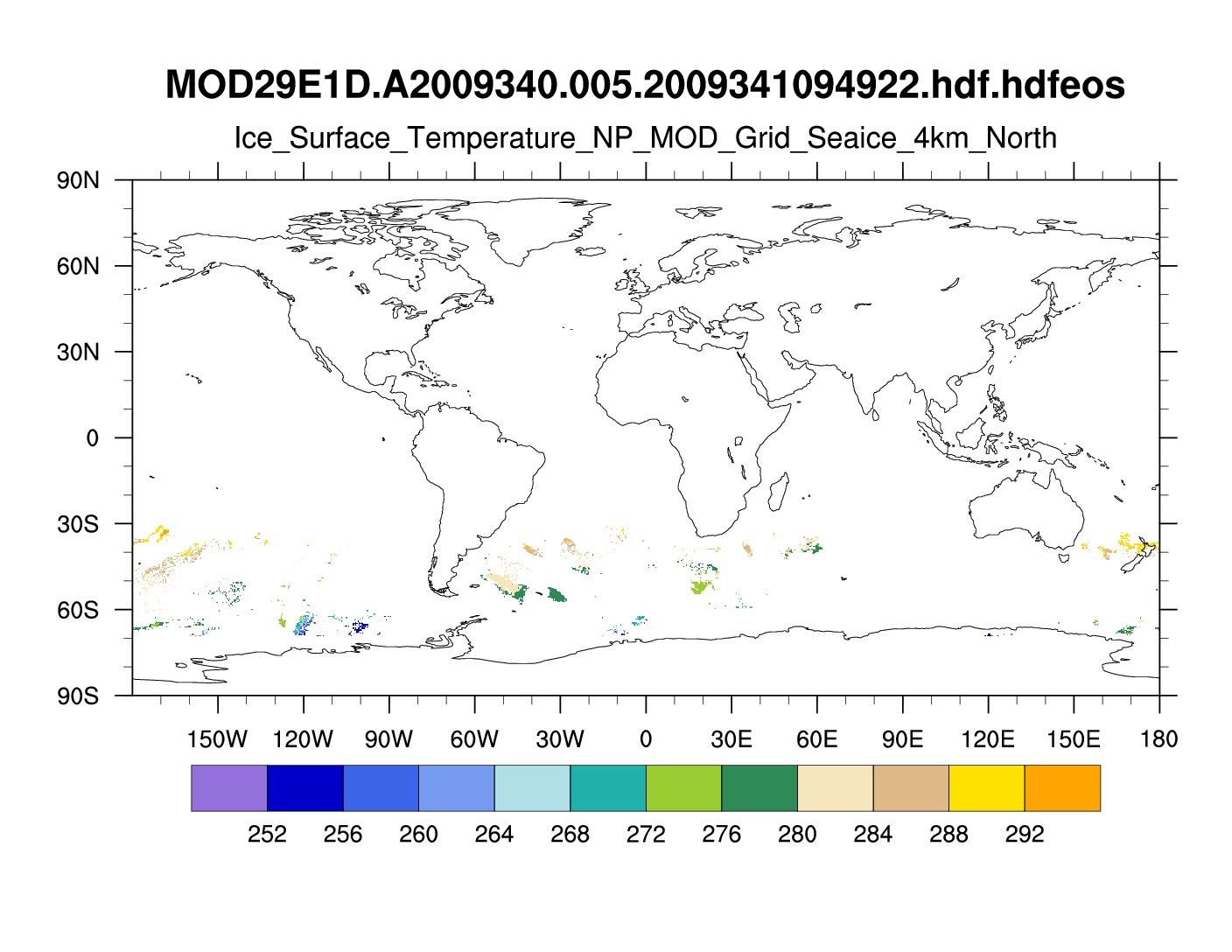
I created the attached ncl script and gif file.
The plotting is 'slow' for this 4501x4501 grid
Good luck
On 04/13/2010 03:01 PM, Tong Qi wrote:
> Hello NCL-Talk,
>
> I am trying to plot the data field "Ice_Surface_Temperature_NP" from
> this hdfeos file:
> ftp://ftp.hdfgroup.uiuc.edu/pub/outgoing/veer/MAP_DATA/files/NSIDC/MODIS/MOD29E1D/MOD29E1D.A2009340.005.2009341094922.hdf
>
> However, when I run my script, I receive:
>
> fatal:NclMalloc Failed:[errno=12]
> Segmentation fault
>
> When I try to use "plot=gsn_csm_contour_map(wks,data,res)" I find that
> the error is generated on that line. Any help is greatly appreciated.
>
> Thanks,
> Tong
>
>
>
> _______________________________________________
> ncl-talk mailing list
> List instructions, subscriber options, unsubscribe:
> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
--
======================================================
Dennis J. Shea tel: 303-497-1361 |
P.O. Box 3000 fax: 303-497-1333 |
Climate Analysis Section |
Climate & Global Dynamics Div. |
National Center for Atmospheric Research |
Boulder, CO 80307 |
USA email: shea 'at' ucar.edu |
======================================================
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk
- text/plain attachment: hdfeos_tong.ncl_MOD29E1D_1d