Date: Fri Jul 15 2011 - 10:01:12 MDT
Dear,
I've tried to make "country mask data" using shapefile and "gc_inout"
function.
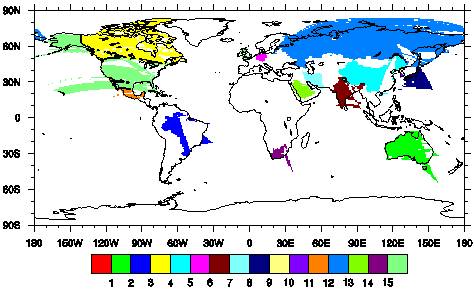
There was no error, but figure looks awkward(The figure is attached.).
I cannot find out what the problem is...
I downloaded shapefile from http://www.mappinghacks.com/data
Here is my ncl script.
------------------------------------------------------------------->
cname = (/"Australia","Brazil"/) ;,"Canada","China","Germany"\
; ,"India","Iran (Islamic Republic of)", "Japan"\
; ,"Korea, Democratic People's Republic of"\
; ,"Korea, Republic of","Mexico","Russia"\
; ,"Saudi Arabia","South Africa","United Kingdom"\
; ,"United States"/)
cdsz = dimsizes(cname)
print(cdsz)
;;========================================================
;; DATA
;;========================================================
dir = "/home/brighti/WORK/DATA/ETC/TM_WORLD_BORDERS_SIMPL-0.2/"
fi = "TM_WORLD_BORDERS_SIMPL-0.2.shp"
ddir = "/home/brighti/WORK/DATA/ANAL/RAWDATA/"
dfi = "cru_ts_3_10.1901.2009.tmp.dat.nc"
;; Make 180x360 2d data ----------------------------------
land = new((/180,360/),"integer",-9999)
land!0 = "lat"
land!1 = "lon"
land&lat = ispan(-895,895,10)/10.
land&lon = ispan(5,3595,10)/10.
land&lat@units = "degree_north"
land&lon@units = "degree_east"
;; Read data ---------------------------------------------
data = land
lat1d = land&lat
lon1d = land&lon
dsize=dimsizes(data)
nlat = dsize(0)
nlon = dsize(1)
;; Read shapefile ---------------------------------------
f=addfile(dir+fi,"r")
name = f->NAME
segments = f->segments
geometry = f->geometry
segsDims = dimsizes(segments)
dims = dimsizes(geometry)
;; Read global attributes --------------------------------
geom_segIndex = f@geom_segIndex
geom_numSegs = f@geom_numSegs
segs_xyzIndex = f@segs_xyzIndex
segs_numPnts = f@segs_numPnts
lat = f->y
lon = f->x
;; Find country info. ------------------------------------
do icnm = 0,cdsz-1
iicc = icnm+1
idxnm = "index_"+iicc
i = ind(name.eq.cname(icnm)) ;;; CHECK !!!
print(i+" "+name(i))
startSegment = geometry(i, geom_segIndex)
numSegments = geometry(i, geom_numSegs)
snum = segments(startSegment, segs_xyzIndex)
enum = segments(startSegment+numSegments,segs_xyzIndex)-1
print(snum+" : " +enum)
mrb_lat = lat(snum:enum-20)
mrb_lon = lon(snum:enum-20)
;print(mrb_lat)
;print(mrb_lon)
;---Put data in the areas that we don't want masked.
do ilt=0, nlat-1
do iln=0, nlon-1
if(gc_inout(lat1d(ilt),lon1d(iln),mrb_lat,mrb_lon)) then
data(ilt,iln) = icnm+1
end if
end do
end do
data@$idxnm$ = cname(icnm)
delete(i)
delete(startSegment)
delete(numSegments)
delete(snum)
delete(enum)
delete(mrb_lat)
delete(mrb_lon)
delete(idxnm)
end do
;;========================================================
;; Make data file
;;========================================================
system("/bin/rm -f land_temp.nc")
fout = addfile("land_temp.nc","c")
fout->landmask = data
;------------------------------------------------------------------------------------------------------------------->
Brighti.
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk