Date: Tue Mar 27 2012 - 10:49:07 MDT
On Mar 27, 2012, at 10:09 AM, Jeff Steward wrote:
> Hi Mary,
>
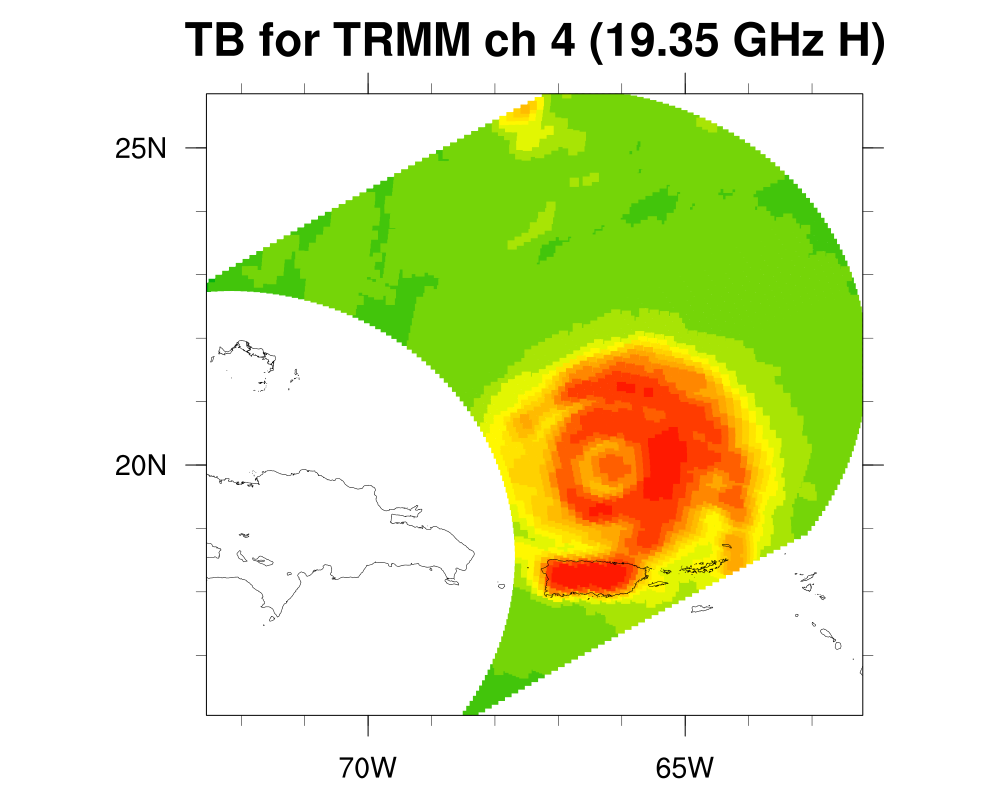
> Thank you. This looks quite a bit better. There is still about half
> of the concave area filled in, but I could probably live with this
> amount if we can't find a better solution.
>
> Alternately I may go to plotting colored scatter data to avoid
> contouring altogether. It won't be as pretty, but it will be more
> accurate. Do you have any suggestions about the best function to use
> for this purpose?
>
> Thank you again very much for your time. Best wishes,
Jeff,
I put together a scatter plot quickly and attached a sample image. You will likely need to change the code to get the colors and levels to your liking.
Also, I used NhlNewMarker to define a filled square as a marker. You can use any one of the characters in our font tables as a marker
(the filled square was font table #35, character "y").
http://www.ncl.ucar.edu/Document/Graphics/font_tables.shtml
Click on any font table line to see all the characters for that table. Or, you can use one of our predefined markers:
http://www.ncl.ucar.edu/Document/Graphics/Images/markers.png
Also, I didn't have time to create a label bar, but there are some examples at
http://www.ncl.ucar.edu/Applications/labelbar.shtml
Example 11 might be the best one.
--Mary
>
> Jeff
>
> On Tue, Mar 27, 2012 at 7:47 AM, Mary Haley <haley@ucar.edu> wrote:
>> Hi Jeff,
>>
>> Thanks for providing your files. Dave will probably look into this further, but one thing I tried was "raster" fill instead of the default "area" fill:
>>
>> opts@cnFillMode = "RasterFill"
>> opts@cnRasterSmoothingOn = True
>>
>>
>> You can try setting cnRasterSmoothingOn to False (the default behavior) to see the "blocky" rasters.
>>
>> BTW, you can use the special "short2flt" function on your raw data to have it automatically apply the scale and offset:
>>
>> val_low_raw = short2flt(raw->lowResCh)
>> val_high_raw = short2flt(raw->highResCh)
>>
>> However, this won't get you the results that your script expects, because "short2flt" actually *multiples* the scale factor rather than divide by it.
>>
>> --Mary
>>
>>
>> On Mar 26, 2012, at 8:27 PM, Jeff Steward wrote:
>>
>>> I have uploaded my file as "plotBT-raw.ncl" and my example HDF file as
>>> "1B11.20100831.72869.7.HDF" under the incoming directory. Thank you.
>>> Best wishes, Jeff
>>>
>>> On Mon, Mar 26, 2012 at 4:57 PM, David Brown <dbrown@ucar.edu> wrote:
>>>> You would need to send us your script and the necessary data (or a link to an accessible data server) for us to help you.
>>>> See the "Report bugs" page under Support on the NCL web site for instructions on uploading scripts and dasta.
>>>> -dave
>>>>
>>>>
>>>> On Mar 26, 2012, at 4:11 PM, Jeff Steward wrote:
>>>>
>>>>> Hello,
>>>>>
>>>>> I need to plot level 1B data (brightness temperatures) from TRMM/TMI,
>>>>> and I have had some measure of success using gsn_csm_contour_map.
>>>>> However, each scan by TRMM is conical, and thus it creates a
>>>>> non-convex path, like this:
>>>>>
>>>>> http://sharaku.eorc.jaxa.jp/ADEOS2/JAXA_TYP_DB/TYP_DB_TRMM/201008/07L/2A12.100829.72843.6.07L.EARL.png
>>>>>
>>>>> However, when I plot this with gsn_csm_contour_map, the concave
>>>>> portion of the scan (the "U" shape at the back of the swath) is filled
>>>>> in with interpolated, very poor quality values; I would much rather
>>>>> just leave those values out. As an example, over land the values
>>>>> should obviously be around 300 K; with the interpolated values, the
>>>>> temperature of the ocean and the land are mixed together. Any ideas
>>>>> about how I can make my plot more like the sharaku link above?
>>>>>
>>>>> Thank you for your time. Best wishes,
>>>>>
>>>>> Jeff
>>>>> _______________________________________________
>>>>> ncl-talk mailing list
>>>>> List instructions, subscriber options, unsubscribe:
>>>>> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>>>>
>>> _______________________________________________
>>> ncl-talk mailing list
>>> List instructions, subscriber options, unsubscribe:
>>> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>>
> _______________________________________________
> ncl-talk mailing list
> List instructions, subscriber options, unsubscribe:
> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk
- application/octet-stream attachment: plotBT-raw-scatter.ncl