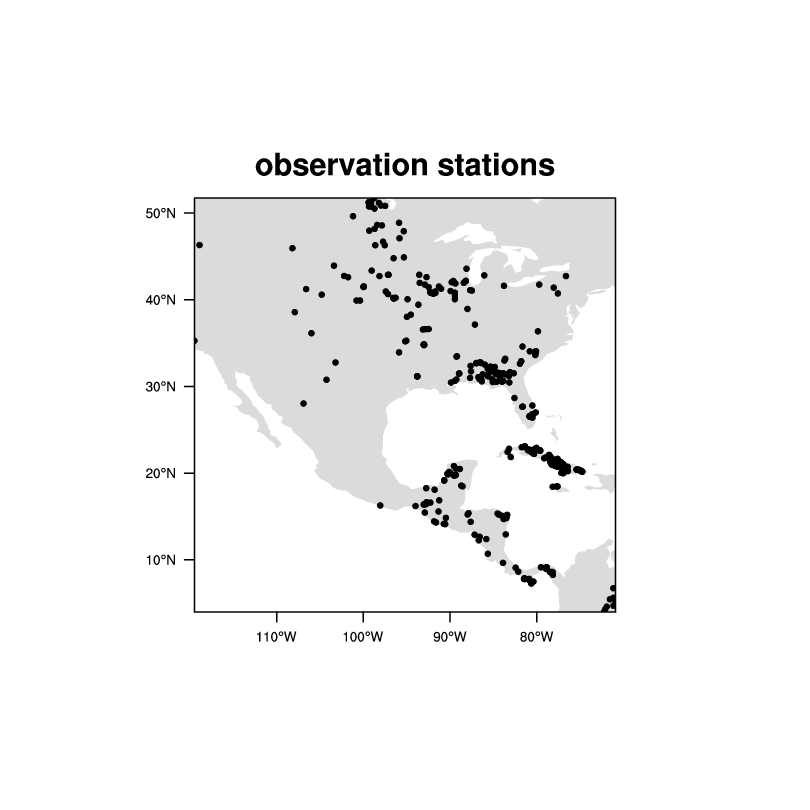
Date: Tue Oct 30 2012 - 14:42:51 MDT
These are not 'pixel level data'. They look like observation
stations. Any interpolated grid would have *highly*
questionable data over most of the area.
See attachment:
On 10/30/12 2:18 PM, Mary Haley wrote:
> Okay, I guess your "x" *doesn't* have coordinate arrays. This looks like unstructured data.
>
> Try adding these two lines:
>
> Opt@SrcGridLat = sfile->latin
> Opt@SrcGridLon = silfe->lonin
>
> You might additionally need to set Opt@SrcMask2D if you have missing values. Something like this:
>
>
> Opt@SrcMask2D = where(.not.ismissing(x),1,0) ; Creates array of 0s and 1s, where 1 represents a non-missing value
>
> --Mary
>
> On Oct 30, 2012, at 2:06 PM, Luo, Chao wrote:
>
>> I have tried this and still there is error. The message is:
>> write_grid_description: can't determine what type of source grid you have.
>> I think this is related to lat/lon of var I want to interpolate. Following is the script I have tried. Please guide me what's wrong of my script. Thanks!
>>
>> CL
>>
>> ---------------------------
>> begin
>>
>> ;---Interpolation methods to use
>> methods = (/"bilinear","patch","conserve"/)
>> nmethods = dimsizes(methods)
>>
>> ;---Read variable to regrid
>>
>> srcFileName = "../dat/biomass_burning_20120313_18_23.nc"
>> sfile = addfile(srcFileName,"r") ; source grid
>>
>> x = sfile->CO_emission ; 6 x 1129
>> var = x(:,0) ; first time
>> latin = sfile->Latitude
>> latin@units = "degrees_north"
>> lonin = sfile->Longitude
>> lonin@units = "degrees_east"
>>
>> dimx = dimsizes( var )
>> nlat = dimx(0)
>> mlon = dimx(0)
>>
>> print(nlat)
>> print(mlon)
>>
>> ;---Create destination lat/lon arrays
>> NLAT36 = 112
>> MLON36 = 148
>>
>> ; lat36 = new (/NLAT36,MLON36/)
>> ; lon36 = new (/NLAT36,MLON36/)
>>
>> lat36 = asciiread("/data11/cluo/cmaq4.6/data/post/obs/grids_148x112/crogrids_lat_148x112.dat" \
>> , (/NLAT36,MLON36/),"float")
>> lon36 = asciiread("/data11/cluo/cmaq4.6/data/post/obs/grids_148x112/crogrids_lon_148x112.dat" \
>> , (/NLAT36,MLON36/),"float")
>>
>> Opt = True ; Regridding options
>> Opt@ForceOverwrite = True
>> Opt@DstGridLat = lat36
>> Opt@DstGridLon = lon36
>>
>> var_regrid = ESMF_regrid(var,Opt)
>>
>> -----------------------
>>
>> ----- Original Message -----
>> From: "Mary Haley" <haley@ucar.edu>
>> To: "Chao Luo" <chao.luo@eas.gatech.edu>
>> Cc: "ncl-talk Talk" <ncl-talk@ucar.edu>
>> Sent: Tuesday, October 30, 2012 10:50:05 AM
>> Subject: Re: interpolate pixel level data into regular grid
>>
>> Chao,
>>
>> Dennis suggested the ESMF regridding routines.
>>
>> You might want to look at ESMF_regrid specifically.
>>
>> Since you have 1D coordinate arrays for the source grid, and want a 12 km x 12 km destination grid, I suggest looking at example ESMF_regrid_2.ncl:
>>
>> http://www.ncl.ucar.edu/Applications/ESMF.shtml
>>
>> This shows how to regrid from a rectilinear grid (which it looks like you have), to a lat/lon grid that you define, which I assume you have.
>>
>> This example uses all three methods of interpolation (bilinear, patch, conserve) and regrids to two different grids.
>>
>> I think you just need to set:
>>
>> ---Common resources
>> Opt = True
>>
>> Opt@ForceOverwrite = True
>>
>> Opt@DstGridLat = lat12
>> Opt@DstGridLon = lon12
>>
>> where lat12 and lon12 contain arrays that represent the 12 km grid.
>>
>> Then, call ESMF_regrid:
>>
>> var_regrid = ESMF_regrid(var,Opt)
>>
>> Where "var" is the variable you want to regrid that contains the 1D coordinate arrays.
>>
>>
>> --Mary
>>
>>
>>
>> On Oct 30, 2012, at 11:09 AM, Luo, Chao wrote:
>>
>>> x refer to time. This is hourly biomass emission data from 18:00-23:00 on 3/17/2012. What I want to do is to interpolate this pixel level data to regular grid (12km x 12km), but don't know which function can be used. Any suggestions are very appreciated!
>>>
>>> ----- Original Message -----
>>> From: "Dennis Shea" <shea@ucar.edu>
>>> To: "Chao Luo" <chao.luo@eas.gatech.edu>
>>> Cc: "ncl-talk" <ncl-talk@ucar.edu>
>>> Sent: Monday, October 29, 2012 5:00:34 PM
>>> Subject: Re: interpolate pixel level data into regular grid
>>>
>>> The issue is "What does the named dimension 'x' refer to?"
>>> There is nothing on the file to indicate its purpose.
>>> You must find out before proceding.
>>>
>>> Attached is a simple NCL script that plots the data.
>>>
>>> netcdf biomass_burning_20120317_18_23 {
>>> dimensions:
>>> x = 6 ;
>>> y = 371 ;
>>> variables:
>>> float Latitude(y) ;
>>> Latitude:long_name = "Latitude" ;
>>> Latitude:units = "degree" ;
>>> Latitude:valid_range = -90.f, 90.f ;
>>> Latitude:scale_factor = 1.f ;
>>> Latitude:add_offset = 0.f ;
>>> Latitude:_FillValue = -9.f ;
>>> float Longitude(y) ;
>>> Longitude:long_name = "Longitude" ;
>>> Longitude:units = "degree" ;
>>> Longitude:valid_range = -180.f, 180.f ;
>>> Longitude:scale_factor = 1.f ;
>>> Longitude:add_offset = 0.f ;
>>> Longitude:_FillValue = -9.f ;
>>> float Burned_area(y, x) ;
>>> [snip]
>>>
>>> On 10/29/12 5:15 PM, Luo, Chao wrote:
>>>> Hi Dennis,
>>>>
>>>> I have checked example 8 and 15. They both require two dimensional lat/lon. The lat/lon of files I have are 1 dimension, but data array is 2D (I am not familiar with these format data). Do I need convert lat/lon from 1D to 2D, and then do interpolation in this case? I am attaching the file. Any suggestions and help are very appreciated!
>>>>
>>>> CL
>>>>
>>>> ----- Original Message -----
>>>> From: "Dennis Shea" <shea@ucar.edu>
>>>> To: "Chao Luo" <chao.luo@eas.gatech.edu>
>>>> Cc: "ncl-talk" <ncl-talk@ucar.edu>
>>>> Sent: Monday, October 29, 2012 3:35:54 PM
>>>> Subject: Re: interpolate pixel level data into regular grid
>>>>
>>>> http://www.ncl.ucar.edu/Applications/ESMF.shtml
>>>>
>>>> Examples 8 and 15 may be of use.
>>>>
>>>> On 10/29/12 3:59 PM, Luo, Chao wrote:
>>>>> Hi,
>>>>>
>>>>> I was wondering if NCL has some functions, which can be used to
>>>>> interpolate satellite pixel level data into regular data. Any helps are
>>>>> very appreciated.
>>>>>
>>>>> CL
>>>>>
>>>>>
>>>>> _______________________________________________
>>>>> ncl-talk mailing list
>>>>> List instructions, subscriber options, unsubscribe:
>>>>> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>>>>>
>>> _______________________________________________
>>> ncl-talk mailing list
>>> List instructions, subscriber options, unsubscribe:
>>> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>> _______________________________________________
>> ncl-talk mailing list
>> List instructions, subscriber options, unsubscribe:
>> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>
> _______________________________________________
> ncl-talk mailing list
> List instructions, subscriber options, unsubscribe:
> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk