Date: Mon Jan 20 2014 - 02:55:09 MST
hi,
i have met a question when i use shapefile file.
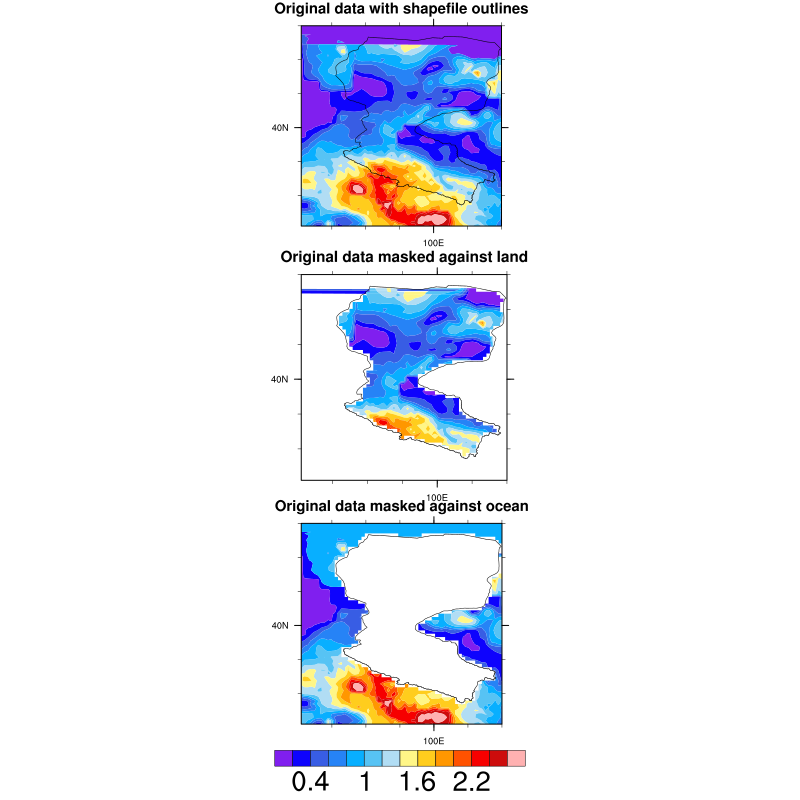
in the middle panel, it's strange that the masked domain exists value, but in the bottom panel, the masked area have not any value.
they have the same frame.
my script used in terms of mask_12.ncl from www.ncl.ucar.edu/Applications/shapefiles.shtml , as follows:
;----------------------------------------------------------------------
; mask_12.ncl
;
; Concepts illustrated:
; - Using a worldwide shapefile to create a land/ocean mask
; - Masking a data array based on a geographical area
; - Attaching shapefile polylines to a map plot
; - Attaching lat/lon points to a map using gsn_coordinates
;----------------------------------------------------------------------
; Downloaded GSHHS shapefiles from:
;
; http://www.ngdc.noaa.gov/mgg/shorelines/data/gshhg/latest/
;
; Used the "coarsest" one: "GSHHS_shp/c/GSHHS_c_L1.shp".
;----------------------------------------------------------------------
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_code.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/gsn_csm.ncl"
load "$NCARG_ROOT/lib/ncarg/nclscripts/csm/contributed.ncl"
;----------------------------------------------------------------------
; Function : create_mask_from_shapefile
;
; This function takes a shapefile of type "polygon" and a 2D data
; array that has coordinate arrays, and creates a mask array
; the same size as the data, that contains 0s and 1s, depending if
; the lat/lon values of the data are inside (1) or outside (0) of the
; shapefile polygons.
;----------------------------------------------------------------------
undef("create_mask_from_shapefile")
function create_mask_from_shapefile(data[*][*]:numeric,fname[1]:string)
local f, segments, geometry, segsDims, geomDims, geom_segIndex, \
geom_numSegs, segs_xyzIndex, segs_numPnts, numFeatures, ilat, ilon, i, j, \
shp_lat, shp_lon, data_lat, data_lon, startSegment, numSegments, seg, \
startPT, endPT, mask_val
begin
;---Error checking
if(.not.isfilepresent(fname)) then
print("Error: create_mask_from_shapefile:")
print(" '" + fname + "' doesn't exist.")
print(" Mask array with all missing values will be returned.")
return(new(dimsizes(data),integer,-999))
end if
;---Check that "data" has 1D coordinate arrays.
if(.not.isdimnamed(data,0).or..not.isdimnamed(data,1).or.\
.not.iscoord(data,data!0).or..not.iscoord(data,data!1)) then
print("Error: create_mask_from_shapefile:")
print(" Input data doesn't have 1D coordinate arrays")
print(" Mask array with all missing values will be returned.")
return(new(dimsizes(data),integer,-999))
end if
;---Open the shapefile
f = addfile(fname,"r")
;---We can't use this routine to mask against point or line data
if(f@geometry_type.ne."polygon") then
print("Error: create_mask_from_shapefile: geometry_type attribute must be 'polygon'")
print(" Mask array with all missing values will be returned.")
return(new(dimsizes(data),integer,-999))
end if
;---Read data off the shapefile
segments = f->segments
geometry = f->geometry
segsDims = dimsizes(segments)
geomDims = dimsizes(geometry)
;---Read global attributes
geom_segIndex = f@geom_segIndex
geom_numSegs = f@geom_numSegs
segs_xyzIndex = f@segs_xyzIndex
segs_numPnts = f@segs_numPnts
numFeatures = geomDims(0)
;---Create mask array
data_mask = new(dimsizes(data),integer,-999)
mask_val = 1 ; 1's represent being inside a polygon
data_mask = 0 ; 0's represent being outside a polygon
;---Read lat/lon values
shp_lon = f->x
shp_lat = f->y
data_lat = data&$data!0$
data_lon = data&$data!1$
nlat = dimsizes(data_lat)
nlon = dimsizes(data_lon)
do i=0, numFeatures-1
startSegment = geometry(i, geom_segIndex)
numSegments = geometry(i, geom_numSegs)
do seg=startSegment, startSegment+numSegments-1
startPT = segments(seg, segs_xyzIndex)
endPT = startPT + segments(seg, segs_numPnts) - 1
;
; Messy code to make the nested loop go faster. Only check
; the area of data that is close to polygon that we are
; traversing. If you check data's full lat/lon array against
; every shapefile polygon, this loop will be extremely slow.
;
;
; Get approx. index values where data's lat/lon is close
; to shapefile's current polygon.
;
iilt_beg = ind(data_lat.le.min(shp_lat(startPT:endPT)))
iilt_end = ind(data_lat.ge.max(shp_lat(startPT:endPT)))
iiln_beg = ind(data_lon.le.min(shp_lon(startPT:endPT)))
iiln_end = ind(data_lon.ge.max(shp_lon(startPT:endPT)))
ilt_beg = 0
iln_beg = 0
ilt_end = nlat-1
iln_end = nlon-1
if(.not.any(ismissing(iilt_beg))) then
ilt_beg = iilt_beg(dimsizes(iilt_beg)-1)
end if
if(.not.any(ismissing(iilt_end))) then
ilt_end = iilt_end(0)
end if
if(.not.any(ismissing(iiln_beg))) then
iln_beg = iiln_beg(dimsizes(iiln_beg)-1)
end if
if(.not.any(ismissing(iiln_end))) then
iln_end = iiln_end(0)
end if
;
; Loop across subset of data's lat/lon and check each point
; to see if it is inside or outside of the shapefile polygon.
;
do ilat = ilt_beg,ilt_end
do ilon = iln_beg,iln_end
if(data_mask(ilat,ilon).ne.mask_val.and.\
gc_inout(data_lat(ilat),data_lon(ilon),\
shp_lat(startPT:endPT),shp_lon(startPT:endPT))) then
data_mask(ilat,ilon) = mask_val
end if
end do
end do
delete([/iilt_beg,iilt_end,iiln_beg,iiln_end/])
end do
end do
return(data_mask)
end
;----------------------------------------------------------------------
; Main code
;----------------------------------------------------------------------
begin
WRITE_MASK = True
DEBUG = False
dir="/public/home/douyj/wks_dyj/clm3.5/bld/clmrun_heihe_0.1d/"
cdf_prefix="clmtest.clm2.h0.2008-08-21-00000"
sfile=dir+cdf_prefix+".nc"
ff=addfile(sfile,"r")
ta=ff->TBOT(0,:,:)
le1=ff->FGEV(0,:,:)
le2=ff->FCEV(0,:,:)
le3=ff->FCTR(0,:,:)
lamda=(2.501-0.00236*(ta-273.15))*1000000
data=(le1+le2+le3)/lamda*24*3600
lat1d=ff->latixy(:,0)
lon1d=ff->longxy(0,:)
lat1d@units="degrees_north"
lon1d@units="degrees_east"
data!0="lat"
data!1="lon"
data&lat=lat1d
data&lon=lon1d
data@_FillValue=-9999
shpfile="/public/home/douyj/wk/heihe_shapefile/heihe_hw.shp"
heihe_mask=create_mask_from_shapefile(data,shpfile)
;---Mask "u" against land and ocean.
u_land_mask = where(heihe_mask.eq.1,data,data@_FillValue)
u_ocean_mask = where(heihe_mask.eq.0,data,data@_FillValue)
copy_VarMeta(data,u_land_mask)
copy_VarMeta(data,u_ocean_mask)
;---Start the graphics
wks = gsn_open_wks("png","mask")
res = True
res@gsnMaximize = True ; maximize plot in frame
res@gsnDraw = False ; don't draw plot yet
res@gsnFrame = False ; don't advance frame yet
res@cnFillOn = True
res@cnLineLabelsOn = False
res@cnLinesOn = False
;---Make sure both plots have same contour levels
mnmxint = nice_mnmxintvl(min(data),max(data),25,False)
res@cnLevelSelectionMode = "ManualLevels"
res@cnMinLevelValF = mnmxint(0)
res@cnMaxLevelValF = mnmxint(1)
res@cnLevelSpacingF = mnmxint(2)
res@lbLabelBarOn = False
res@gsnAddCyclic = False
res@mpFillOn = False
res@mpOutlineOn = False
res@gsnRightString = ""
res@gsnLeftString = ""
res@mpMinLatF = min(lat1d)
res@mpMaxLatF = max(lat1d)
res@mpMinLonF = min(lon1d)
res@mpMaxLonF = max(lon1d)
;---Create plot of original data and attach shapefile outlines
res@tiMainString = "Original data with shapefile outlines"
map_data = gsn_csm_contour_map(wks,data,res)
dum1 = gsn_add_shapefile_polylines(wks,map_data,shpfile,False)
;---Create plots of masked data
res@tiMainString = "Original data masked against land"
map_land_mask = gsn_csm_contour_map(wks,u_land_mask,res)
res@tiMainString = "Original data masked against ocean"
map_ocean_mask = gsn_csm_contour_map(wks,u_ocean_mask,res)
if(DEBUG) then
mkres = True
; mkres@gsMarkerSizeF = 0.007
mkres@gsnCoordsAttach = True
gsn_coordinates(wks,map_data,data,mkres)
mkres@gsnCoordsNonMissingColor = "yellow"
mkres@gsnCoordsMissingColor = "black"
gsn_coordinates(wks,map_land_mask,u_land_mask,mkres)
gsn_coordinates(wks,map_ocean_mask,u_ocean_mask,mkres)
end if
;---Add shapefile outlines
dum2 = gsn_add_shapefile_polylines(wks,map_land_mask,shpfile,False)
dum3 = gsn_add_shapefile_polylines(wks,map_ocean_mask,shpfile,False)
;---Draw all three plots on one page
pres = True
pres@gsnMaximize = True
pres@gsnPanelLabelBar = True
gsn_panel(wks,(/map_data,map_land_mask,map_ocean_mask/),(/3,1/),pres)
if(WRITE_MASK) then
delete(ff) ; Close file before we open again.
;
; Make copy of file so we don't overwrite original.
; This is not necessary, but it's safer.
;
new_cdf_file = cdf_prefix + "_with_mask.nc"
system("/bin/cp " + sfile + " " + new_cdf_file)
finout = addfile(new_cdf_file,"w")
filevardef(finout, "land_mask", typeof(heihe_mask), (/ "lat", "lon" /) )
finout->land_mask = (/heihe_mask/)
end if
end
;--------------------------------------------------------------------------
my question is about how to get rid of the value in my masked area?
thanks
dyjbean@gmail.com
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk