Date: Mon Apr 22 2013 - 13:37:39 MDT
Kamal,
It looks to me like NCL is plotting correctly. Your June and January "oro" values are not the same on both files. Perhaps the oro values on the January file are incorrect?
I used a simple "print" statement to verify that your "oro" values are different on both files:
oro_jan = in_jan->LANDMASK(0,:,:)
oro_jun = in_jun->LANDMASK(0,:,:)
print("Number of January oro=1 values = " + num(oro_jan.eq.1))
print("Number of January oro=0 values = " + num(oro_jan.eq.0))
print("Number of June oro=1 values = " + num(oro_jun.eq.1))
print("Number of June oro=0 values = " + num(oro_jun.eq.0))
The results were:
(0) Number of January oro=1 values = 33178
(0) Number of January oro=0 values = 5798
(0) Number of June oro=1 values = 30044
(0) Number of June oro=0 values = 8932
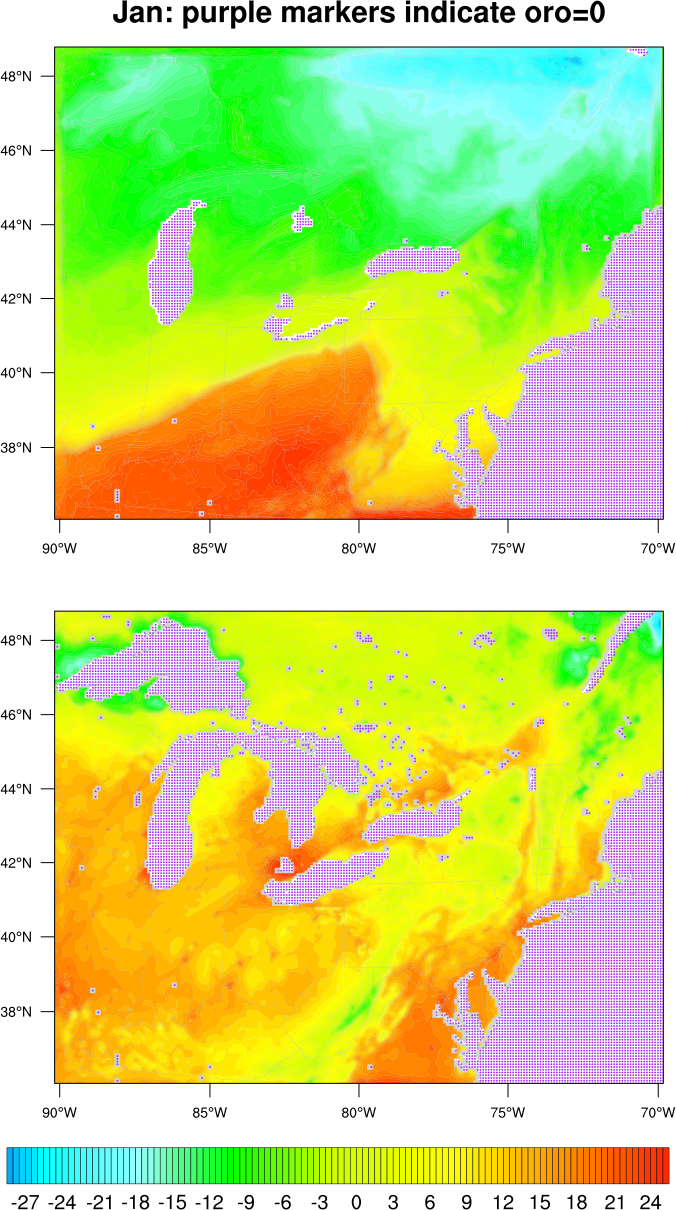
I then added markers to show the locations where "oro.eq.0". See attached script and plot.
--Mary
On Apr 22, 2013, at 11:59 AM, mmkamal@uwaterloo.ca wrote:
> Hi Mary,
>
>
> I have checked the data and find that it is okay. I am sharing the data with you via my dropbox https://www.dropbox.com/home/WRF%20Data . Please let me know if you can download the data or not.
>
>
> Thanks
> Kamal
>
>
>
>
>
> Quoting Mary Haley <haley@ucar.edu>:
>
>> Hi Kamal,
>>
>> Are you sure you are applying the correct mask to the January data?
>>
>> In your script below, you reference index "22" for the ts and oro variables. If this is the time dimension, did you update this index in both locations when you changed from July to January?
>>
>> If you continue to have problems with this script, it will help if you can make the data available.
>>
>> Thanks,
>>
>> --Mary
>>
>> On Apr 18, 2013, at 4:05 PM, mmkamal@uwaterloo.ca wrote:
>>
>>> Hi,
>>>
>>> I was trying to mask out points over water for WRF's T2 and RAINC and RAINNC variables by adjusting NCL's http://www.ncl.ucar.edu/Applications/Scripts/mask_1.ncl script as per Mary Haley's suggestion (http://www.ncl.ucar.edu/Support/talk_archives/2013/1000.html). But I was wondering after looking at the results. It gives me perfect result for the month of July but does incomplete masking in the month of January. I am attaching the resultant images for your convenience. Here is my code:
>>>
>>>
>>> Thanks
>>> Kamal
>>>
>>>
>>> =======================================================
>>>
>>> in=addfile("/scratch/j/jcl/mkamal/wrfout_d02_2002-01-01.nc","r")
>>>
>>> ts = in->T2(22,:,:)-273.15
>>> oro = in->LANDMASK(22,:,:)
>>>
>>> ;************************************************
>>> ; use mask function to mask out land and then ocean data
>>> ; water=0,land=1
>>> ;************************************************
>>> land_only = ts ; trick to keep cv's and atts
>>> ocean_only = ts
>>>
>>> land_only = mask(ts,oro,1) ; Return the values of ts where oro=1
>>> ocean_only = mask(ts,oro,0) ; Return the values of ts where oro=0
>>>
>>> ;************************************************
>>> ; common resources
>>> ;************************************************
>>> wks = gsn_open_wks("png","wrf_d02_temp_mask") ; open a ps file
>>> gsn_define_colormap(wks,"BlAqGrYeOrRe") ; choose colormap
>>>
>>> res = True ; plot mods desired
>>> res@cnFillOn = True ; turn on color fill
>>> res@cnLinesOn = False ; turn of contour lines
>>>
>>> res@gsnSpreadColors = True ; use full colormap
>>> res@gsnSpreadColorStart = 10 ; start at color 10
>>> res@gsnSpreadColorEnd = 96 ; end at color 96
>>>
>>> ;************************************************
>>> ; Use WRF_contributed procedure to set map resources
>>> ;************************************************
>>> WRF_map_c(in, res, 0) ; reads info from file
>>>
>>> ;************************************************
>>> ; if appropriate, set True for native (direct) mapping (faster)
>>> ;************************************************
>>> res@tfDoNDCOverlay = True
>>>
>>> ;************************************************
>>> ; associate the 2-dimensional coordinates to the variable for plotting
>>> ;************************************************
>>> if (.not.res@tfDoNDCOverlay) then
>>> lat2d = f->XLAT(0,:,:)
>>> lon2d = f->XLONG(0,:,:)
>>>
>>> ocean_only@lat2d = lat2d
>>> ocean_only@lon2d = lon2d
>>> land_only@lat2d = lat2d
>>> land_only@lon2d = lon2d
>>> end if
>>>
>>> ;************************************************
>>> ; NCL contouring does not like _FillValue=0.0
>>> ;************************************************
>>> land_only@_FillValue = -999. ; change to -999
>>> ocean_only@_FillValue = -999.
>>>
>>>
>>> ;************************************************
>>> ; individual plots
>>> ;************************************************
>>> res@cnLevelSpacingF = 0.5 ; interval
>>> res@lbLabelStride = 1 ; every 4th label
>>> res@tiMainString = "WRF Land Only" ; title
>>> plot = gsn_csm_contour_map(wks,land_only,res)
>>>
>>> res@cnLevelSpacingF = 0.5 ; interval
>>> res@lbLabelStride = 1 ; every 2nd label
>>> res@tiMainString = "WRF Ocean Only" ; title
>>> plot = gsn_csm_contour_map(wks,ocean_only,res)
>>> end
>>> ===================================================
>>>
>>>
>>>
>>>
>>> <July_T2_Land.png><July_T2_Ocean.png><January_T2_Land.png><January_T2_Ocean.png>_______________________________________________
>>> ncl-talk mailing list
>>> List instructions, subscriber options, unsubscribe:
>>> http://mailman.ucar.edu/mailman/listinfo/ncl-talk
>>
>>
>
>
>
>
_______________________________________________
ncl-talk mailing list
List instructions, subscriber options, unsubscribe:
http://mailman.ucar.edu/mailman/listinfo/ncl-talk
- application/octet-stream attachment: wrf.ncl